Section: New Results
Computational Physiology
Modeling and Simulation of Longitudinal Brain MRIs with Atrophy in Alzheimer's Disease
Participants : Bishesh Khanal [Correspondant] , Nicholas Ayache, Xavier Pennec.
Alzheimer's Disease (AD), modeling atrophy, bio-physical model, simulation
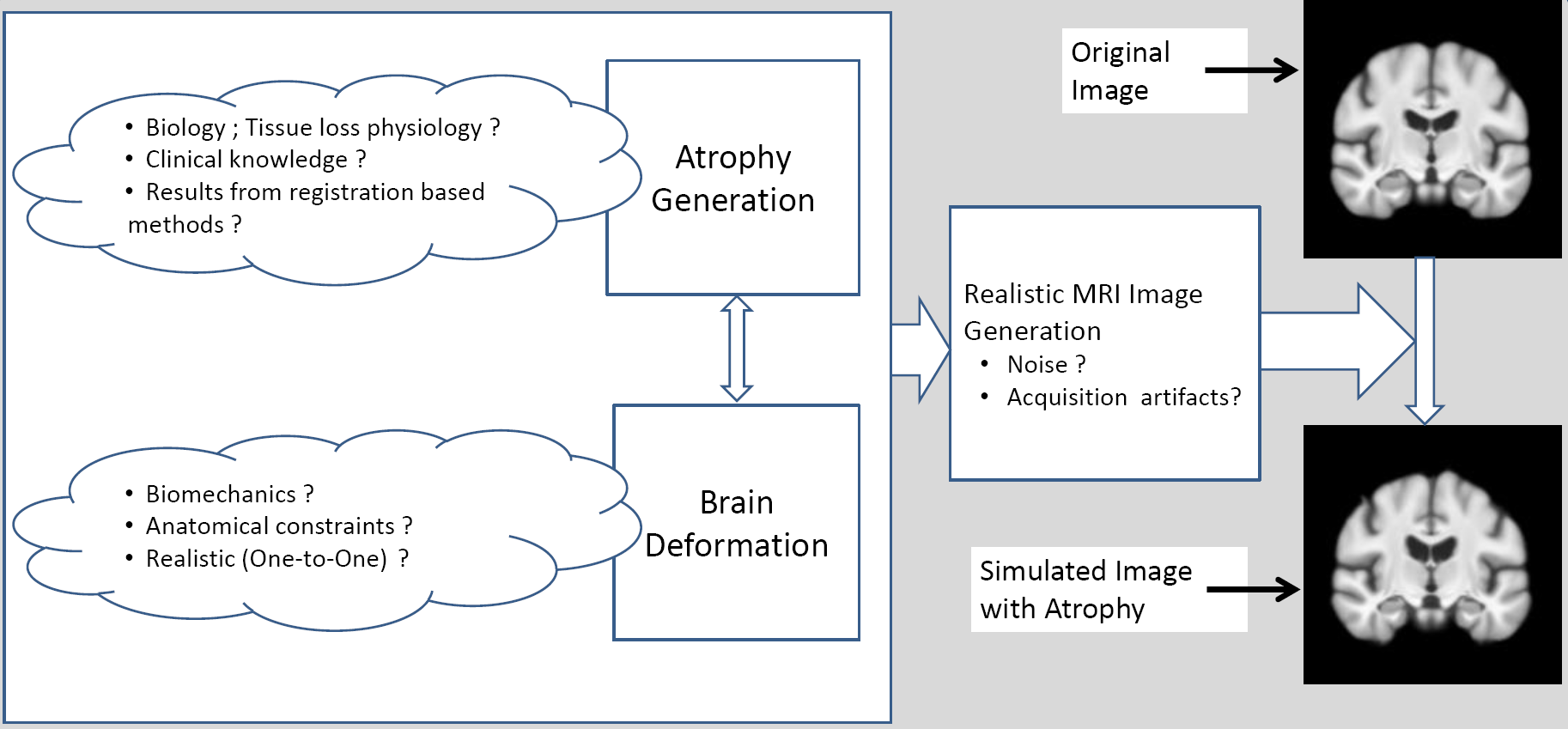
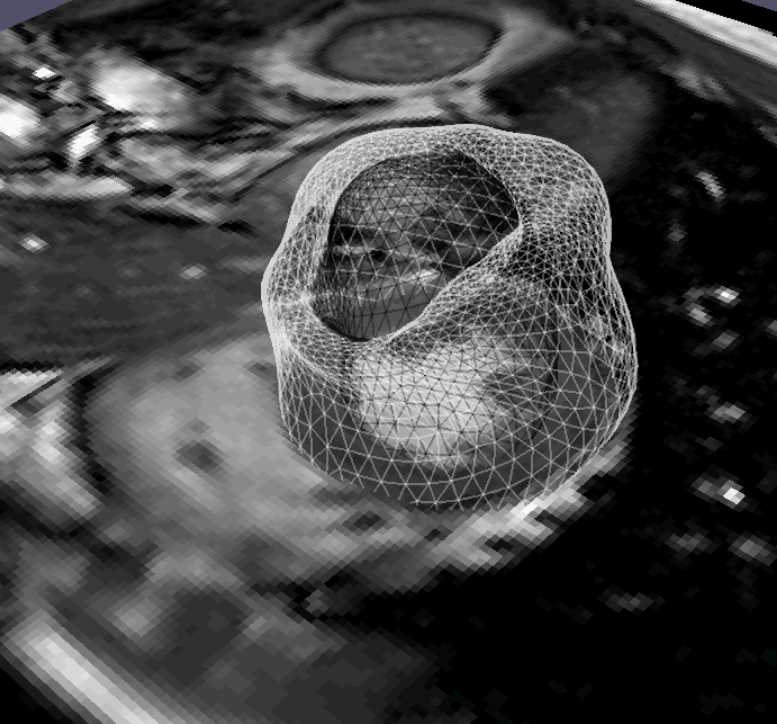
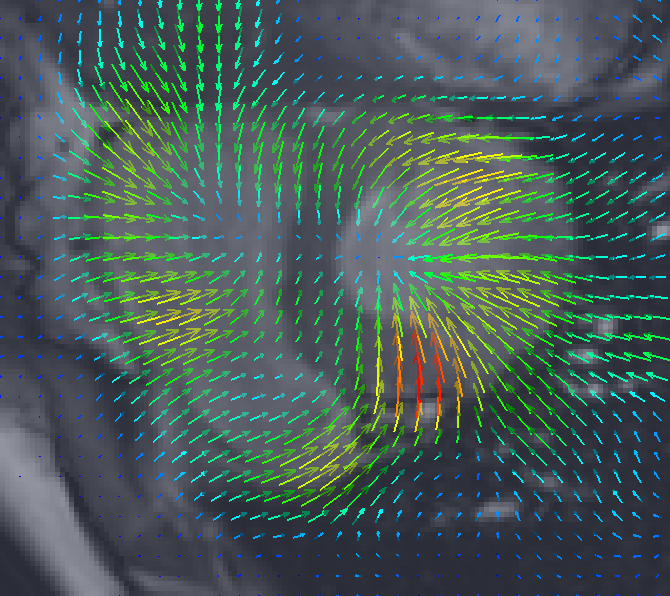
We have implemented a 3D bio-physical model for the deformation of the brain with Alzheimer's Disease (AD).The model produces a deformation field of the brain when a known distribution of local volume change (atrophy) is given as the input. The obtained deformation is then used to warp the original 3D MR image. The major contribution of this work corresponds to the block “Brain Deformation” in Figure 7 .
Registration of time series of cardiac images
Participants : Loic Le Folgoc [Correspondant] , Hervé Delingette, Antonio Criminisi, Nicholas Ayache.
This work has been partly supported by Microsoft Research through its PhD Scholarship Programme and the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Registration, Automatic Relevance Determination, Magnetic resonance, 3D-US
-
We developed a generic approach to registration building on the framework of Automatic Relevance Determination. We applied this framework to the tracking of heart motion throughout time series of images from cine-MR, tagged-MR and 3D-US modalities.
-
Our approach allows for the joint determination of model parameters, such as noise and regularization parameters, decreasing the need for manual tuning and preprocessing. Moreover, it is suitable for further analysis of uncertainty in the output of the registration.
|
Real-Time Cardiac Electrophysiology Computing for Training Simulator
Participants : Hugo Talbot [Correspondant] , Hervé Delingette, Stephane Cotin, Maxime Sermesant, Christian Duriez.
This work was performed in collaboration with the SHACRA team in Lille.
Cardiac electrophysiology simulation, Cryoablation simulation, SOFA framework, GPU computing, patient-specific study
-
Cardiac arrhythmia is a very frequent pathology that comes from abnormal electrical activity in the myocardium. This work aims at developing a training simulator for cardiologists in the context of catheterization and thermo-ablation of these arrhythmias. After tackling the issue of fast electrophysiology computation [27] , a first version of our training simulator was proposed which combines virtual catheterization and interactive GPU electrophysiology modeling [70] . This year, the simulator has been improved by tackling the issue of interactive catheter navigation inside a moving venous system and a beating heart [70] . The simulator was demonstrated during the VRIPHYS 2013 workshop in Lille and the Inria-industry meeting in Paris. Personalization of the electrophysiological model using the data assimilation library Verdandi has been initiated.
-
Cryotherapy simulation in collaboration with the IHU Strasbourg has been performed. This technique consists in inserting needles that freeze the surrounding tissues, thus immediately leading to cellular death of the tissues. We built a simulator able to place the cryoprobes and run a simulation representing the evolution of iceballs in living tissues [58] .
Personalized model of the heart for cardiac therapy planning
Participants : Stéphanie Marchesseau [Correspondant] , Maxime Sermesant, Hervé Delingette, Nicholas Ayache.
This work is performed in the context of the PhD of Stéphanie Marchesseau in collaboration with St Thomas Hospital in London and was partially funded by ERC MedYMA.
-
Personalization of the mechanical function of the heart from time series of cardiac images has been achieved by combining global calibration of a few global parameters [18] with estimation of regional contractility parameters [17] using data assimilation techniques.
-
Personalized cardiac models were used to create synthetic images [22] of cardiac motion thus allowing the benchmarking of motion tracking algorithms [8] , [39] .
Cardiac Arrhythmia Radio-frequency Ablation Planning
Participants : Rocio Cabrera Lozoya [Correspondant] , Maxime Sermesant, Hervé Delingette, Nicholas Ayache.
This work is performed in the context of the PhD of Rocío Cabrera Lozoya in collaboration with the CHU LIRYC Bordeaux and is funded by ERC MedYMA.
-
Biophysical model development for the prediction of radio frequency ablation sites for ventricular tachycardias.
-
Structural and functional characterization of target sites using 3D imaging and EP measurements through machine learning algorithms.
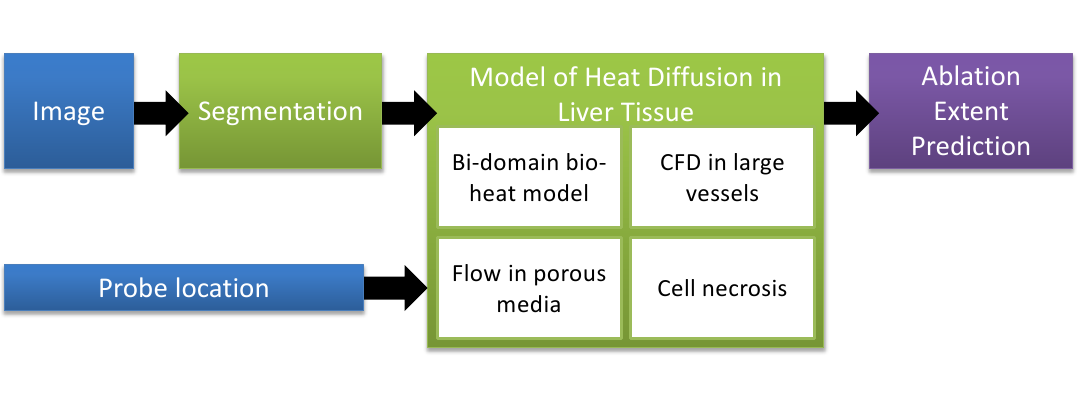
Computational modeling of radiofrequency ablation for the planning and guidance of abdominal tumor treatment
Participants : Chloé Audigier [Correspondant] , Hervé Delingette, Tommaso Mansi, Nicholas Ayache.
This PhD is carried out jointly between the Asclepios research group, Inria Sophia Antipolis, France and the Image Analytics and Informatics global field, Siemens Corporate Research, Princeton, USA.
Radio Frequency Abation, Patient-Specific Simulation, Lattice Boltzmann Method, Computational Fluid Dynamics, Heat Transfer, Therapy Planning, Liver
-
In order to obtain a computational framework for patient-specific planning of radiofrequency ablation, a patient-specific detailed anatomical model of the liver has been extracted from a standard CT image and then meshed with tetrahedra. The structures of interest include : parenchyma, lesion, hepatic vein and vena cava.
-
A computational fluid dynamic model is used to estimate the patient-specific blood flow in the hepatic circulatory system. It was combined with a porous media model to compute the patient-specific blood flow distribution inside the parenchyma using the results of the CFD solver (pressures).
-
Bio-heat equations and a cell death model to account for cellular necrosis have been implemented with FEM using SOFA and a Lattice Boltmann Model to model heat propagation in biological tissues [35] leading to improved accuracy and computational efficiency.
Tumor growth assessment based on biophysical modeling
Participants : Erin Stretton [Correspondant] , Bjoern Menze, Nicholas Ayache, Hervé Delingette.
This work was carried out during the Phd of Erin Stretton and was funded by the Care4Me project. It was performed in collaboration with Pr Mandonnet, Lariboisière hospital in Paris, and the German Cancer Research Center (DKFZ)
Glioma simulation, tumor growth.
We aim at developing image analysis methods [23] using tumor growth models in order to guide the planning of therapies (surgical removal and chemotherapy) for brain cancer (glioma) patients. Our work is focused on these objectives :
-
Predicting the location of glioma recurrence after a resection surgery;
-
Determining the description the of tumor cell diffusion tensor in white matter (patient-based, atlas based or isotropic) which leads to the most accurate results for predicting future tumor growth [57] ;
-
Comparing tumor growth speeds between 8 patient cases based on biophysical modeling and various manual methods.
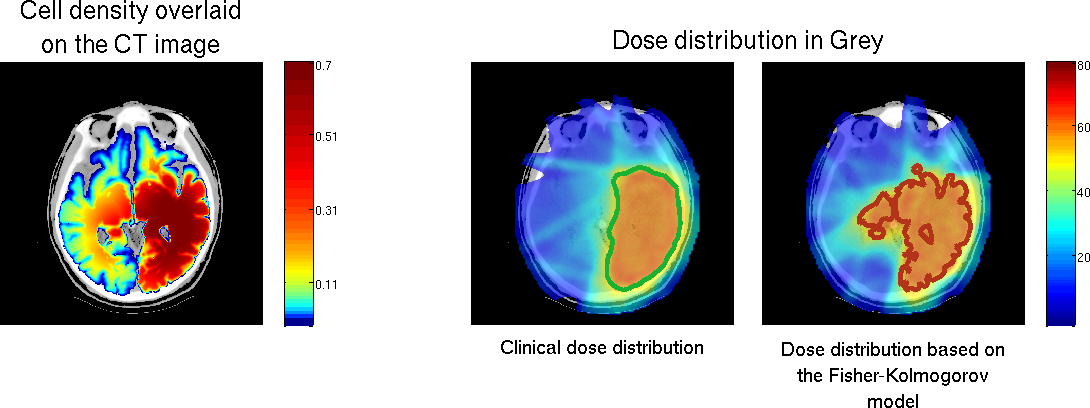
Brain tumor growth modeling : Application to radiation therapy
Participants : Matthieu Lê [Correspondant] , Jan Unkelbach, Nicholas Ayache, Hervé Delingette.
This work is carried out between Asclepios research group, and the Department of Radiation Oncology of the Massachusetts General Hospital, Boston, USA. Part of this work was funded by the European Research Council through the ERC Advanced Grant MedYMA.
Glioma simulations, radiation therapy, target delineation, vasogenic edema
-
We developed a tumor growth model for high grade gliomas, based on different types of cell and the vascularization of the brain.
-
We studied multimodal brain tumor images to evaluate tumor infiltration.
-
We used a Fisher-Kolmogorov model to improve target volume delineation for radiation therapy (see Figure 11 )
|
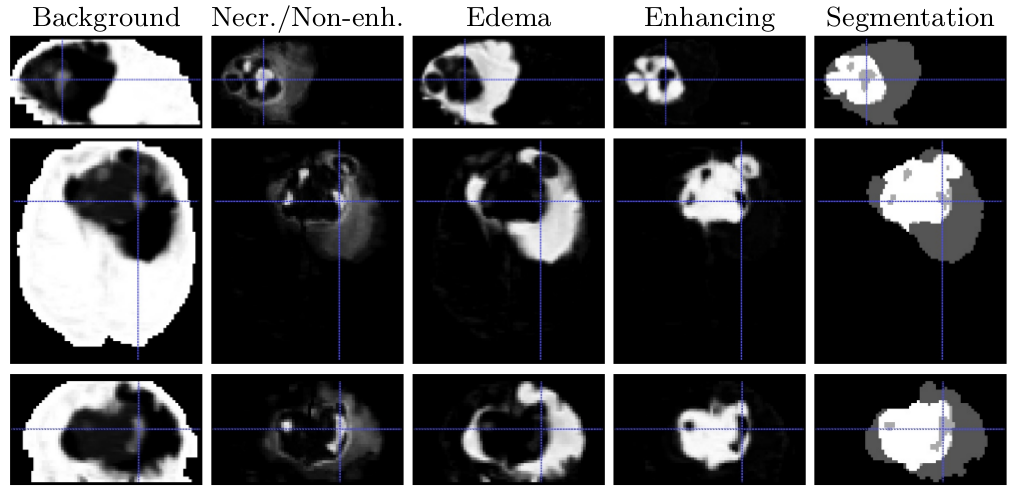
Multimodal patch-based glioma segmentation
Participants : Nicolas Cordier [Correspondant] , Bjoern Menze, Hervé Delingette, Nicholas Ayache.
Part of this work was funded by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain, MRI, Glioma, Patch-based Segmentation, Tumor Simulation
-
A patch-based approach was developed for glioma segmentation based on multi-channel 3D MRI. The method is fully automatic and does not require any learning step.
-
Features: multi-channel MR intensities in local neighborhoods.
-
A heuristic label fusion strategy was introduced and showed promising results, as shown in Figure 12 .
-
The algorithm was ranked 5th in the Brain Tumor Segmentation Challenge (BraTS) at MICCAI 2013 [67] .
-
Large unlabeled glioma MRI databases are being incorporated in the framework.